LABORATORIES
Cell Lineages and Neuronal Heterogeneity laboratory
Cell Lineages and Neuronal Heterogeneity laboratory
Research
Team
Publications
Contact
Others
Research
The brain is one of the organs with the greatest cellular diversity, containing thousands of different types of neurons and glia. Each of these types performs different functions, for which cells have specialized during evolution. This causes them to differ in morphology, gene expression, electrical properties, etc.
The brain of any organism is generated from a single cell, the zygote. This cell divides to ultimately give rise to all the cells that make up an organism. Cell division must be perfectly coordinated with the cell specification process by which cells acquire a specific cellular identity. Cell types generated at different times or from different progenitors must therefore acquire a precise identity.
Our laboratory tries to understand the cellular specification processes that are behind the generation of the nervous system. In addition, we also want to emulate these processes to generate cell types of therapeutic interest.
Tools
Star Track
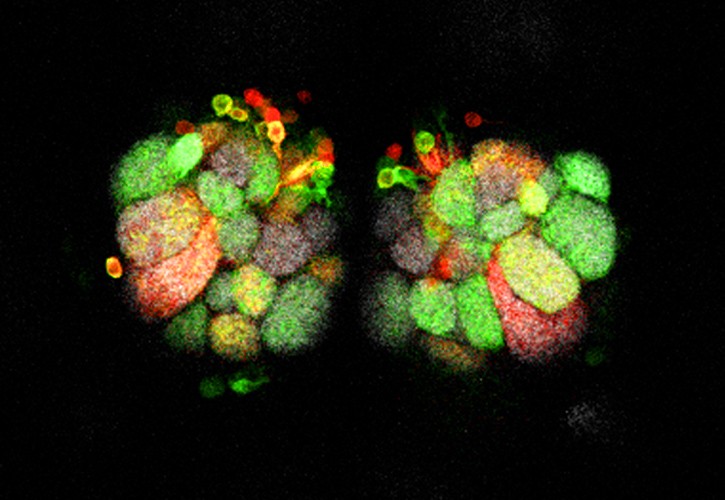
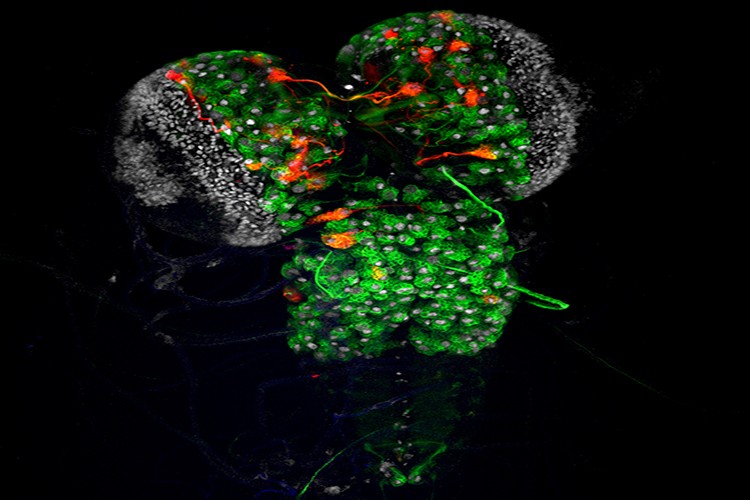
Designed to study mouse astrocyte lineages, Star Track makes it possible to label different clones of astrocytes or NG2 cells with a specific combination of fluorescent proteins. This method is based on the in utero electroporation of a mixture of 13 plasmids. This mixture is made up of 6 plasmids, each with a GFAP promoter (astrocyte marker with weak expression in NG2 cells) that regulates the cytoplasmic expression of a fluorescent protein, and another 6 for nuclear expression. Because the tag that is generated has to be stable and not diluted with cell divisions, these plasmids also contain the Piggybac transposon sequences. In addition, the plasmid mixture also includes a plasmid that ubiquitously encodes the Piggybac transposase and promotes the integration of the other plasmids.

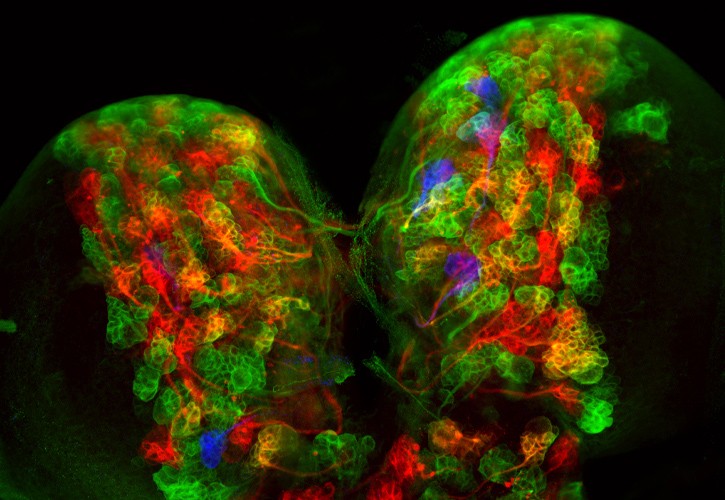
Mouse olfactory bulb marked with Star Track
After the mixture of the 13 plasmids is electroporated into the embryonic or postnatal ventricle, the progenitors surrounding the ventricle acquire most of the plasmids. However, by chance, only some of these plasmids will integrate and therefore the parent will contain a specific combination of all 12 plasmids. Due to integration into the genome mediated by the Piggybac transposon, all progeny derived from the same parent will inherit the same combination of fluorescent proteins. In this way, clones of astrocytes or NG2 cells will show a unique color in the cytoplasm and nucleus, which serves to resolve the different lineages.
CaSSA
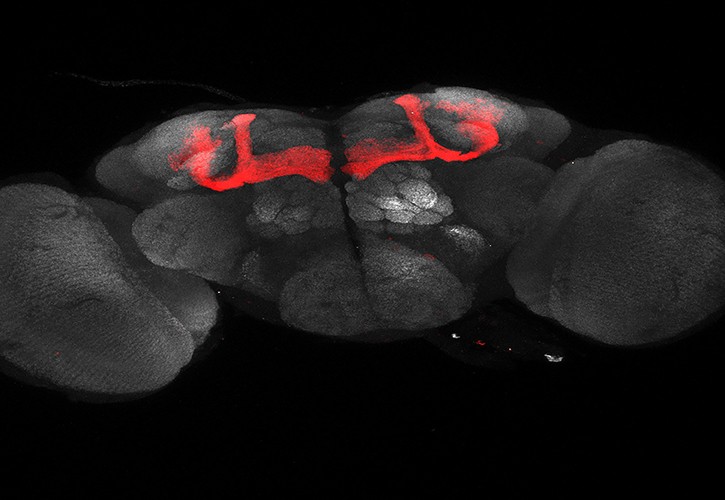
Different cell types can be defined based on the expression of a specific combination of marker genes. Therefore, if one wants to gain genetic access to a specific cell type, one of the routes involves directing the genetic manipulation to those cells that express a combination of genetic markers through methods of genetic intersection. The problem is that these methods usually only get an intersection of two genetic markers.
CaSSA emerges as an alternative to classical genetic intersections with recombinases or other molecular biology tools. This tool is based on two essential elements: 1) CRISPR/Cas9, an endonuclease that cuts DNA in that region of 20 base pairs encoded by a gRNA, and 2) Single Strand Annealing (SSA), an evolutionarily conserved mechanism for repair. of DNA repeat regions. In CaSSA, the gene to be expressed (for example, reporters such as GFP or effectors such as a recombinase), is interrupted by several activation regions. These regions consist of two direct repeats of a part of the gene, separated by one or multiple sequences recognized by a specific gRNA. Because gRNA encodes a 20 base pair sequence, an almost unlimited number of specific gRNAs and thus distinct activation regions can be designed. When a specific gRNA recognizes the activating region in the presence of Cas9, this enzyme cuts the DNA. This cut induces repair by SSA, which results in the elimination of one of the repeats and of the region recognized by the gRNA, thus reconstituting the original sequence of the gene to be expressed in that region.
CLADES
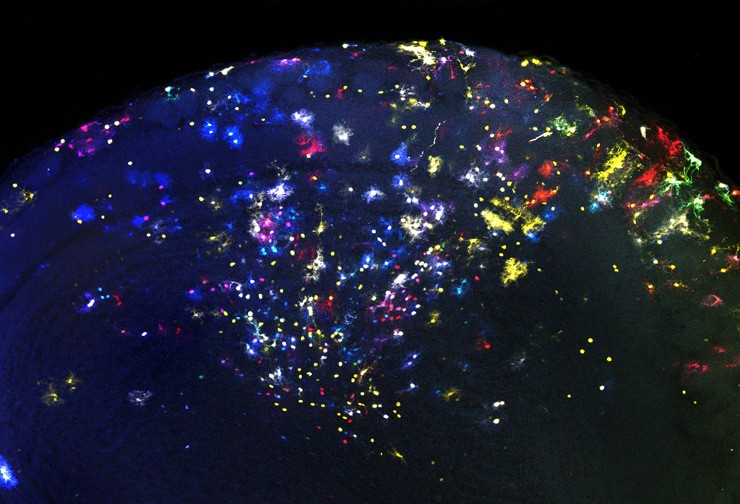
Until now, most lineage analysis work has involved marking clones of cells from the same parent. However, this analysis does not allow us to see the structure of the lineage tree, nor to know which cells have been generated in the first divisions and which in the last ones. There are alternatives that allow us to see the structure of the lineage, such as certain tools that trace the spontaneous or induced mutations that occur in DNA throughout development. However, these tools destroy the structure and morphology of the cells, do not allow to follow the lineage with high resolution and have a very high cost.
Until now, most lineage analysis work has involved marking clones of cells from the same parent. However, this analysis does not allow us to see the structure of the lineage tree, nor to know which cells have been generated in the first divisions and which in the last ones. There are alternatives that allow us to see the structure of the lineage, such as certain tools that trace the spontaneous or induced mutations that occur in DNA throughout development. However, these tools destroy the structure and morphology of the cells, do not allow to follow the lineage with high resolution and have a very high cost.
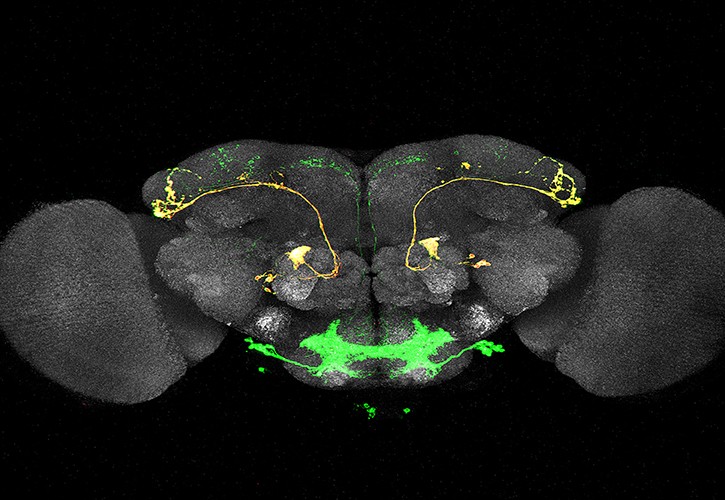
This tool has been optimized in Drosophila and has made it possible to confirm the order in which neurons of certain specific lineages are produced. Its operation is so simple that it will allow you to trace lineages at the single cell level in a short time. This will also make it possible to analyze phenotypes of mutants in which the lineage is altered.
Team

Jorge García-Marqués
Principal Investigator
Want to join our team?
On the occasion of our upcoming addition to the Cajal Institute, we are currently recruiting new members for the group. If you are interested, contact us at:
Publications
Publicaciones de los últimos años
- A programmable sequence of reporters for lineage analysis
Garcia-Marques J.*, Espinosa-Medina, I., Ku, K., Yang, C., Koyama, M., Yu, H., Lee, T.
*Co-autor de correspondencia
NATURE NEUROSCIENCE 23:1618-1628 (2020)
DOI: 10.1101/655308 - Unlimited genetic switches for cell-type-specific manipulation
Garcia-Marques J, Yang CP, Espinosa-Medina I, Mok K, Koyama M, Lee T.
NEURON (2019)
DOI: 10.1016/j.neuron.2019.07.005 - NG2-glia from pallial progenitors produce the largest clonal clusters of the brain: time frame of clonal generation in Cortex and Olfactory Bulb
Garcia-Marques J, Lopez-Mascaraque L.
J NEUROSCI 34: 2305-2313(2014)
DOI: doi: 10.1523/JNEUROSCI.3060-13.2014 - Clonal identity determines astrocyte cortical heterogeneity
Garcia-Marques J, Lopez-Mascaraque L.
CEREB CORTEX 23: 1463-1472 (2013)
DOI: 10.1093/cercor/bhs134
Contacto
Where to find us
Laboratorio de Linajes celulares y heterogeneidad neuronal. Jorge García Marqués
Instituto Cajal CSIC. Avda. Doctor Arce, 37. 28002. Madrid
Call us
Phone:
Write us
Email address:
Otros

Neuroscience Research Center dependent on the CSIC. Founded in 1920 and initially directed by Santiago Ramón y Cajal. World reference in the study of the brain. Custodian of the Cajal Legacy.
Activities
