WILD CARD OF WANDS
POLYMERASE CHAIN REACTION
Wild card of Wands: PCR or POLYMERASE CHAIN REACTION, a genetic technique that copies the cellular mechanisms of nucleic acid replication
POLYMERASE CHAIN REACTION, or PCR, was a technique unknown to non-specialists in biochemistry or biomedicine until the SARS CoV-2 epidemic put it on everyone’s lips. But PCR is not only useful to know whether or not we are infected by the coronavirus that causes COVID-19, because it is a very powerful technique. Thanks to the thermocycler, which allows the magic of the work of the DNA polymerase enzyme, PCR denatures, hybridizes and elongates to replicate the minimum fragment of genetic material of the organism that we want to study.
The polymerase chain reaction, or PCR by its English acronym, is a technique used in molecular biology that allows a large number of copies of a DNA fragment to be obtained in a short time, starting from a tiny amount of this biomolecule.
DNA is one of the two types of nucleic acid that exist. Nucleic acids are the name given to the biomolecules that store the genetic coding of any living organism: they are the “instruction manual” where, with a code of five bases (adenosine, cytosine, guanosine, thymine and uracil) they are “written” and they specify the characteristics that the living being will have. They can be of two types:
- Double-stranded deoxyribonucleic acid (DNA) [1] and characteristic of animals (including humans), plants, bacteria and some viruses (such as hepatitis B or human papilloma).
- Single-stranded ribonucleic acid (RNA) characteristic of certain viruses (such as the famous SARS CoV-2 that causes COVID-19, among many others)
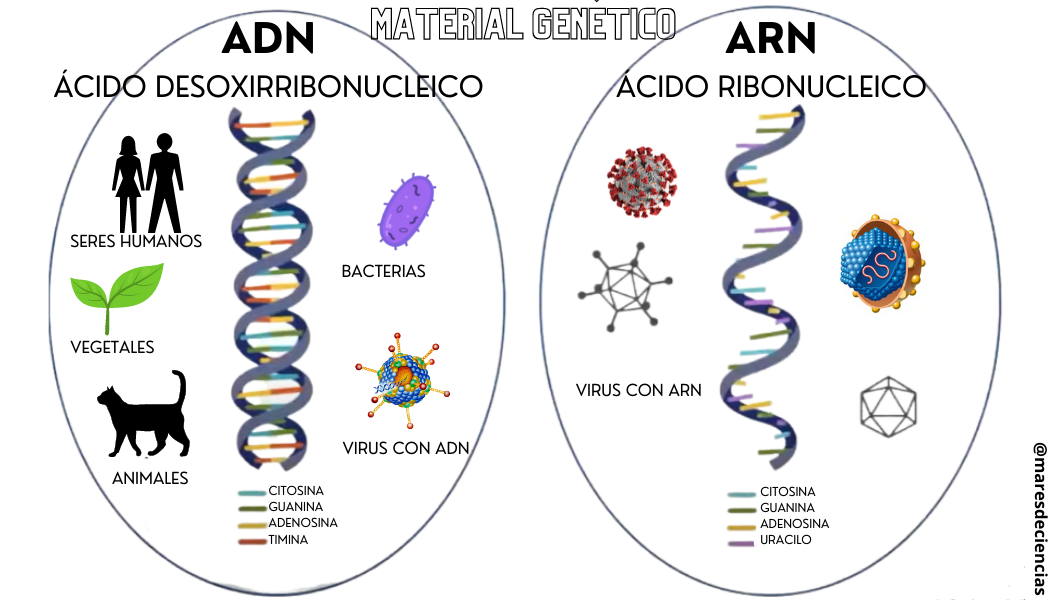
Adapted from https://www.youtube.com/watch?v=xWrxSV83b1g
When the cells of a living organism divide, their genetic material must duplicate so that each daughter cell inherits an identical copy of the parent cell’s genetic material. This process is called mitosis.
To replicate the genetic material and obtain two identical copies, living beings rely on the enzymatic activity of very particular molecules: DNA polymerases. The PCR technique is based on this enzymatic activity that occurs naturally in living cells. As occurs in vivo, in vitro thermostable DNA polymerases (resistant to high temperatures) are used capable of “photocopying” a DNA fragment of which we wish to obtain a large number of equal copies.
Description of the technique:
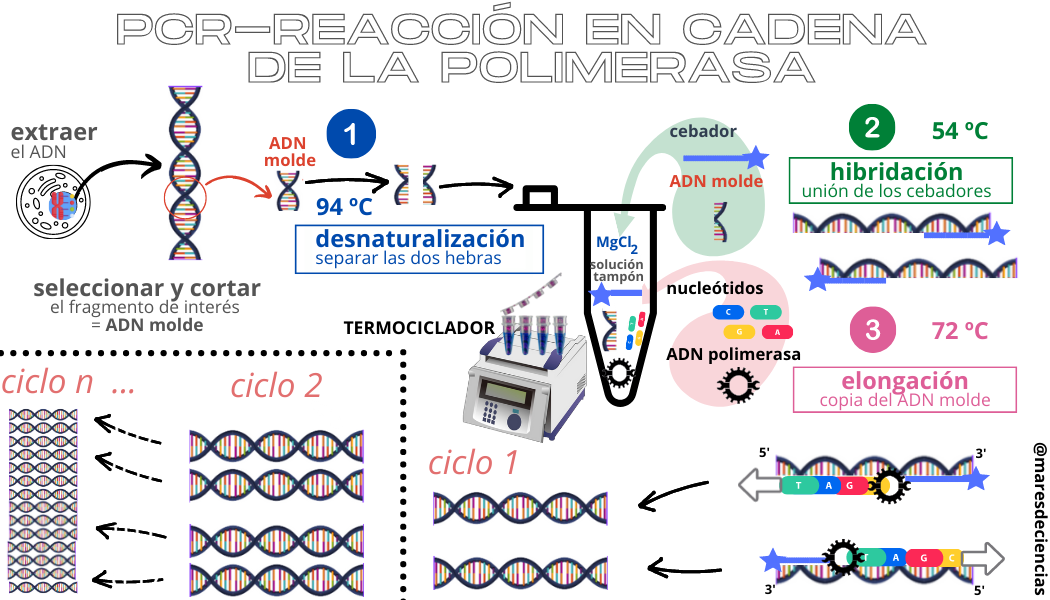
PCR consists of the repetition of an integrative cycle of three stages, each of which is characterized by a specific temperature and duration.
Therefore, to carry out a PCR it is essential to perfectly regulate the temperature conditions in which the successive stages will occur and to precisely establish the duration of each of them. These are the functions of the THERMOCYCLER, which, thanks to the possibility of programming the modification of the temperature of the solution on several occasions and their times, allows this technique to be carried out in much less time, since to carry it out, the temperature of the solution must be modified. solution on several occasions.
In addition to the thermocycler, to carry out a PCR we need the following elements:
- The template DNA or DNA fragment that we want to amplify using PCR.
- Nucleotides (deoxyribonucleotides-triphosphate): These are the four types of nitrogenous bases that form the framework of DNA (adenine, guanine, citosine and thymine).
- Primers (in English, primers). These are oligonucleotides (short DNA sequences with between 15 and 30 nucleotides) that bind to the template DNA molecule and serve as a starting point for Taq polymerase to begin DNA synthesis. In PCR we need two primers, each complementary to a strand of the DNA that we seek to amplify (at the 3′ end). They are the ones that will delimit the region of the DNA that is going to be amplified.
- DNA polymerases – the queens of this biochemical technique. Different DNA polymerases, obtained from various organisms, can be used in PCR. However, the most effective and resistant to high temperatures comes from a bacteria and is called Taq Polymerase.
- Divalent magnesium ions. They are used as cofactors of the polymerase. These cations are essential for the function of DNA polymerase. Normally, magnesium chloride (MgCl2) is added so that magnesium (with a +2 charge) is released when it dissociates.
A buffer solution capable of regulating the pH, that is, the acidity or basicity conditions, of our PCR. This is very important, because changes in the pH of the solution can alter the results of our PCR or prevent it from occurring.
The stages of each cycle are:
DENATURALIZATION. Between 90 ºC and 95 ºC the bonds that create the nitrogenous bases between the two chains are broken and the two strands can be separated.
HYBRIDIZATION. Lowering the temperature to between 50ºC and 65ºC allows the single strands of DNA to aggregate with the extra genetic material (the primers). The objective is to enclose the DNA sequence that you want to amplify between two primers.
ELONGATION. Raising the temperature again between 72 ºC and 75 ºC activates the DNA polymerase and, thanks to the primers, this enzyme can begin to reconstruct the complementary strands and copy only the sequences between two primers. At the end of the cycle, for each single strand a double strand is obtained, so that in each cycle the number of DNA sequences is doubled.
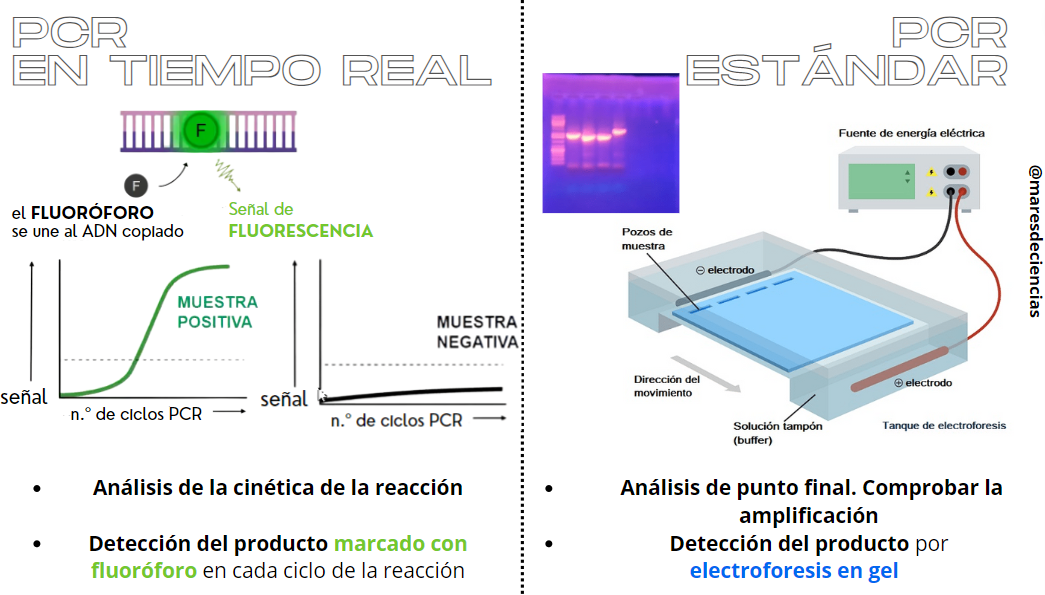
There is a variant of PCR, real-time PCR, which allows the rate of generation of one or more specific amplification products to be measured in a thermocycler provided with fluorescence sensors. In this case, the accumulation of amplified DNA is detected and quantified as the reaction progresses, that is, “in real time.” To do this, it is necessary to add a fluorescent molecule that is associated with the amplified DNA, so that the increase in this fluorescence is proportional to the increase in the amount of amplified DNA molecules in the reaction [6].
The story of a discovery that allowed the birth of genomics
The PCR technique was invented in 1985 by the biochemist Kary B. Mullis, who in 1993 received the Nobel Prize in Chemistry for this discovery (shared with Michael Smith for their work on directed mutagenesis procedures).
Mullis’ idea was to make millions of copies of a DNA fragment quickly and easily, but this required finding a DNA polymerase capable of resisting the high temperatures (94ºC) that were necessary to unfold the strands of the double helix of the DNA. DNA. It was two of Mullis’ colleagues, Susanne Stoffel and David H. Gelfand, who found it in the bacterium Thermus aquaticus that had been discovered by microbiologist Thomas Brock. Another microbiologist, Randy Siki, demonstrated that this Taq DNA polymerase was ideal for replicating DNA in the PCR process, because it could withstand high temperatures without losing its functionality [3] [4]. PCR is a technique that repeats in a loop a cycle of denaturation processes (separation of the DNA chains), hybridization with primers (nucleotide sequences that allow the action of the Taq polymerase to start) and elongation, so that in each cycle the number of DNA copies doubles. In 31 cycles, the following are generated:
2,147,483,648 copies!!!!!
But what is PCR used for?
At this point you are surely thinking why we want to have so many copies of a fragment of genetic material from a cell, a bacteria or a virus. Well, PCR is a star technique for any researcher who works with DNA molecules. Did you know that today it is the technique that allows us to obtain large quantities of synthetic insulin? This is achieved by retrotranscribing the insulin-specific messenger RNA and amplifying it with PCR and then cloning it into an expression vector that allows this protein to be produced in large quantities [5].
The polymerase chain reaction has become popular in colloquial language as a result of the COVID-19 pandemic, but its use in laboratories has been very common for many years, for a wide variety of applications, ranging from DNA sequencing to the generation of forensic DNA profiles from genetic samples, through the identification of pathogens thanks to the detection of the absence or presence of their genes in the infected organism.
Among other things, it allows for the preparation of DNA fragments for cloning into bacterial plasmids or viruses to use them as vectors, an essential step in the development of gene therapies. Furthermore, many clinical diagnoses no longer require long processes of bacterial cultures and biochemical tests, because it is now faster and easier to amplify specific genetic sequences thanks to PCR. The benefits of genetic amplification extend to the diagnosis of hundreds of hereditary diseases or to identify pathogens (such as the coronavirus that causes COVID-19). It is also a very important technique in paternity tests and in forensic analyzes by scientific police.
At the Cajal Institute, the Molecular and Cellular Biology Unit (BMC) provides both support to researchers, especially in matters of genotyping and real-time PCR, and technical support to get the most out of the equipment and materials available. of neuroscientists.

Surely you have been left wanting to know more. By clicking here you can see a video from our colleagues at the Center for Molecular Biology (CBM), a joint CSIC-Autonomous University of Madrid center. They prepared it for Science Week 2021 and it is a good example of the practical application of PCR to the detection of diseases in humans with the case of tests for COVID-19.
Have you been curious and want to know more?
NOTES AND SOURCES
[1] There are some viruses whose DNA is not double-stranded, but rather single-stranded. These viruses make the missing copy to reconstruct the DNA double helix thanks to the cellular machinery of the cells they infect.
[2] PCR only works with DNA fragments. So, for the cases of organisms whose genetic material is made up of RNA, as is the case of viruses with RNA, it is necessary to add a prior step to the technique: RETROTRANSCRIPTION, which allows making a copy of double-stranded DNA from of the single strand of the RNA fragment. And it will be with this copy of DNA that the PCR can be carried out.
[3] SADURNÍ, J. M. (2020). An eccentric Nobel. Kary Banks Mullis, the story of the creator of PCR. History, National Geographic https://historia.nationalgeographic.com.es/a/kary-banks-mullis-eccentricidad-genio_15910
[4] CERNA CORTES, J., CERNA CORTES, J. F., GUAPILLO VARGAS, M.R.B. (2004). Taq polymerase. From geysers to science. Science and Technology Topics vol. 18 number 54 September – December 2014 pp. 52-57. pdf
[5] GARCÍA, RB., MONTES, HM., RAMOS, RE., ARIZA. CA., PÉREZ, VJ., GÓMEZ, GO., CALVA, CG. (2010). Cloning of the cDNA of the human insulin gene in aerial roots of Brassica oleracea var italica (broccoli). CENIC Magazine. Biological Sciences. Vol. 41. 1-9 https://doaj.org/article/33b5d8a6c69a43808c18e0d6e3f4f616
[6] Real-time PCR. Faculty of Chemistry of the National Autonomous University of Mexico https://quimica.unam.mx/investigacion/servicios-para-la-investigacion/usaii/pcr-en-tiempo-real/
OTHER VIDEOS
Eugenia, from the Mitochondria Channel, explains what PCR consists of and the role of the thermocycler https://www.youtube.com/watch?v=NZ5XNjOBptc&t=571s
Crisal, from Canal Un Professor. https://youtu.be/4WdgrcOUn84?t=91
Nutrimente Channel: PCR: polymerase chain reaction (theoretical and practical) https://www.youtube.com/watch?v=mpRy8CSAISs (see up to minute 05:41)
Carlos Cordón, from the CSIC Institute of Molecular Biology, explains the PCR + control electrophoresis process on the We Sapiens Channel https://www.youtube.com/watch?v=whJTMyyV7gU
